Research
3D histopathology using light sheet microscopy
Current gold-standard histopathology for cancerous biopsies is destructive, time consuming, and limited to 2D slices, which does not faithfully represent true 3D tumor micro-morphology. Our research goal is to utilize dual-inverted Selective Plane Illumination Microscopy (diSPIM) for 3D imaging of tumors and rendering their digital histological images. By looking at the 3D volume with less sampling issues caused by physical cutting, and with the help of further computational analysis, the imaging system shows the potential for better understanding the structures of tumors, speeding up triage, and improving diagnosis. Furthermore, by adding electronic confocal slit detection (eCSD) and structured illumination (SI), diSPIM has been optimized to achieve higher image contrast (Figure A), enabling useful pseudo-H&E imaging of even highly-scattering (uncleared) tissues. Also, to improve imaging depth and cover larger 3D volume, optical clearing methods, such as TDE and CLARITY, are also being investigated. Traditional histology-like digital sections can be obtained from the reconstructed 3D volume (Figure B). By utilizing dual-view imaging and joint deconvolution, isotropic resolution over large fields of view can be achieved.
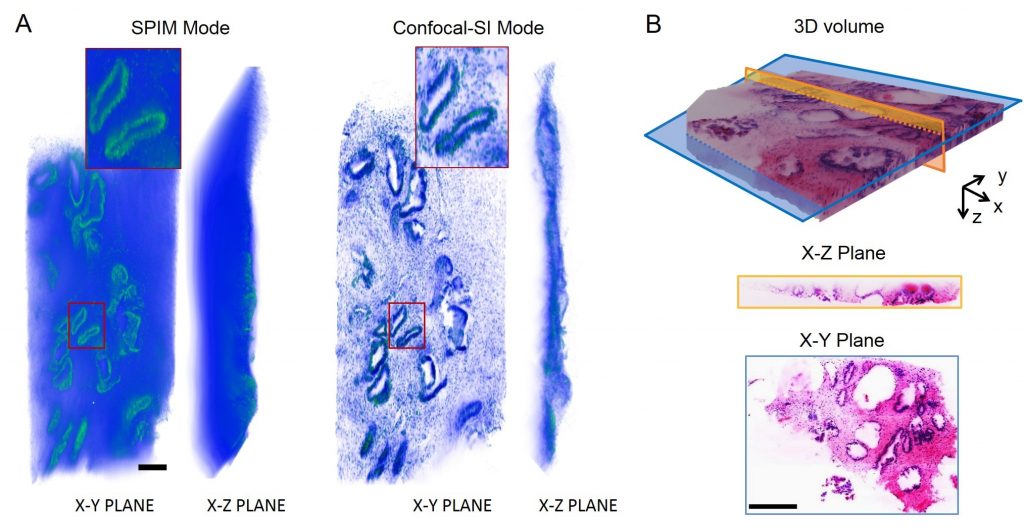
For 3D imaging of uncleared tissue, scattering background deteriorates imaging performance (Fig. A, SPIM Mode). By adding electronic confocal slit detection (eCSD, Confocal mode) and structured illumination (SI mode), imaging contrast has been tremendously improved (Fig. A, Confocal-SI Mode). As for cleared tissue with less scattering background, by imaging and reconstructing D&E stained sample at SPIM mode, traditional histology-like images can be visualized as a 3D volume or sections at arbitrary orientations (Fig. B). Scale bar: 250µm (A), 500µm (B).
Panoramic imaging of whole tumor resection surfaces
1.6 GP panorama of uncut lateral surface of a fresh radical prostatectomy specimen
The frontline curative strategy for prostate cancer is surgical removal of the entire organ via radical prostatectomy. Achieving cancer free surgical margins during surgical treatment for prostate cancer is critical to prevent tumor recurrence and reduce the need for costly adjuvant procedures. The positive surgical margin rate can exceed 50% for more advanced prostate cancers. Frozen section analysis can be utilized to evaluate only a small portion of the margin, therefore intraoperative surgical margin evaluation is limited by time-consuming, labor-intensive, and tissue-destructive processing steps. To address these limitations, we are using structured illumination microscopy (SIM) to rapidly image the surface of excised organs for point-of-care margin evaluation. Current projects include clinical studies utilizing SIM to image freshly excised prostate and kidney surgical margins for intraoperative margin evaluation. In these projects, we also incorporate 3D scanning of the whole organ in order to develop a clinical atlas matching features observed on VR-SIM to 3D anatomy and traditionally stained tissue slides. Our method will allow pathologists to view SIM images with pathologically relevant contrast in subcellular detail in an intuitive 3D space using telepathology.
Rapid on-site biopsy adequacy evaluation
210 MP image of fresh uncut prostate biopsy with HGPIN
Every year, more than 5 million patients in the U.S. undergo biopsy procedures to get definitive cancer diagnoses. The goal of the biopsy procedure is to get a sample that is representative of the suspected cancerous region. However, often patients need repeat procedures because the biopsies were not representative of the suspected area. Many biopsy procedures incorporate a rapid on-site evaluation (ROSE) technique for quality assurance of the sample using touch-imprint cytology. However, the current ROSE does not give a clear picture of the quality of biopsy because the technique only allows evaluation of less than 1% of the cells that have “fallen off” of the biopsy. As a result, ROSE is inaccurate much of the time, leading to costly and painful repeat procedures. Furthermore, ROSE depletes DNA content from the biopsies, impacting downstream molecular analysis and genetic sequencing. We use structured illumination microscopy (SIM) to digitally images biopsy samples without destruction at subcellular resolution within seconds of removal, using simple fluorescent stains. SIM provides an exact picture of the intact biopsy, providing improved ROSE accuracy over current methods. Current projects are focused on using SIM to image renal, prostate, and lung biopsies for cancer evaluation and liver biopsies for transplant evaluation.
under construction….
(With Carola Wenk, Tulane Computer Science, and Brittany Fasy, Montana State University Computer Science)
under construction….
under construction….
Funding for our work is provided by:
National Institutes of Health, National Cancer Institute, Innovative Molecular Analysis Technologies, 1R33CA196457-01
National Institutes of Health, National Cancer Institute, 1R21CA159936-01

National Science Foundation, DMS-1664848
National Science Foundation, IIP-1650948
National Science Foundation, DMS-1557750

